- Biography
- Works
- Contact
The information below refers to the time of the award.
The biologist Venki Ramakrishnan was born in India and is an American citizen. He has worked in the UK since 1999 and is currently the joint Head of the Structural Studies Division at the Medical Research Council’s Laboratory of Molecular Biology in Cambridge. He is a Fellow of the Royal Society and of the US National Academy of Sciences and a member of the European Molecular Biology Organisation.
Studies on the ribosome and therapeutic prospects
Found within cells, the ribosomes are factories for the production for proteins, which are essential for all living organisms to function.
Appearing very early during the evolution of species, ribosomes are present in both micro-organisms and human cells, but in slightly different forms. These differences are exploited by antibiotics, which block bacterial ribosomes without affecting their human counterparts.
Venki Ramakrishnan and his team have obtained images of the atomic structure of the ribosome, i.e. how its atoms are arranged in space. This allowed them to obtain detailed knowledge of the mechanisms of protein translation and, more specifically, to identify the sites where antibiotics bind. They were thus able to obtain a better understanding of how these drugs affect the activity of the ribosome.
This highly basic research is also of great practical importance for medicine. Better targeting of the bacterial ribosome should make it possible to avoid effects on the human ribosome and thus to decrease the secondary effects linked to antibiotic use. It should also make it possible to combat the development of bacterial resistance (for example, resistance to anti-tuberculosis drugs).
From the ribosome ro bacterial resistance to antibiotics
The cellular machinery
Located inside the cells, ribosomes are essential components of the cellular machinery. Indeed, it is in the ribosome that DNA is “translated” into a chain of amino acids, which form the protein.
Genes can be thought of as long sentences made up of words of three letters chosen from a four-letter alphabet consisting of the “bases”, adenine, cytosine, guanine, and thymine (or A, C, G, and T), whereas proteins use a completely different alphabet because they consist of assemblies of about 20 amino acids. The genetic instructions must therefore be “translated” for a protein to be synthesised. This operation is carried out in the ribosome: it is here that the gene, after having been copied into messenger ribonucleic acid (mRNA), is translated by means of transfer RNA (tRNA) into amino acids, the linking of which results in the formation of the protein (see figure 1).
Ribosomes are made up of two subunits, a small one, called “30S” and a larger one, called “50S”, in which the proteins are produced. Together, these two subunits form the complete, or “70S”, molecule.
The ribosome and antibiotics
Formed themselves from long chains of RNA and several proteins, ribosomes appeared very early in evolution. They probably originated in very primitive forms of life in which the genome was made up of RNA, rather than DNA.
Bacterial ribosomes and those of more highly evolved organisms are not completely identical. These differences are exploited therapeutically, as many antibiotics block bacterial ribosomes without affecting their human counterparts.
In order to improve the action of antibiotics, it is essential to understand how ribosomes work and how antibacterial drugs bind to them. In this area, the high-resolution images obtained by Venki Ramakrishnan have led to important advances.
High-resolution images
In 2000, Venki Ramakrishnan and his team described the atomic structure of the small subunit of the bacterial ribosome, i.e., how its atoms are arranged in space. To do this, they irradiated crystals of the 30S subunit with powerful beams of X-rays produced by synchrotrons. They then used programmes which used the scattering of the X-rays caused by the crystals to reconstruct the 3-D image of the subunit.
Using this procedure, the researchers were able to visualise how antibiotics bind to the 30S subunit. They found that the drugs bound to specific sites on the ribosome and blocked its activity (figure 2). They also demonstrated that the ribosome ensures a correct match between the tRNA and the mRNA, improving the accuracy of protein synthesis. By interfering with this process, antibiotics can induce the ribosome to make mistakes.
Forging ahead, Venki RAMAKRISHNAN and his colleagues determined the high-resolution structure of the entire bacterial ribosome in 2006 and were thus able to describe the interactions between it and the different RNAs (messenger and transfer) (figure 3). They also explained how the two subunits (30S and 50S) associate, partly with the help of magnesium ions.
Antibiotics rehabilitated ?
At the basic level, the studies of Venki Ramakrishnan have provided very useful information about the mechanisms governing protein synthesis. However, they could also have interesting therapeutic applications, as the ribosomes are essential elements for the action of antibiotics.
As we all know, the inappropriate prescription of drugs of this type has led to the development of a very worrying phenomenon, as many micro-organisms have become resistant to antibiotics, which have therefore become ineffective. This is especially the case for streptomycin. Long utilised for the treatment of tuberculosis, this antibiotic is now unusable, not only because of its side-effects, but also because the bacillus has become resistant to its action. Since streptomycin acts on the ribosome, we can hope that a better understanding of this particle might allow the situation to be reversed. This is one example, among many, of the potential therapeutic impact of the studies of Venki Ramakrishnan.
The biologist intends to pursue his studies on the ribosome. He will use the Louis-Jeantet Prize to trap the particle in different stages of the process of protein synthesis in order to determine the different forms which it adopts at each of these steps. He also aims to determine the structure of ribosomes from higher organisms, in which the initial step in protein synthesis is far more complex than in bacteria.
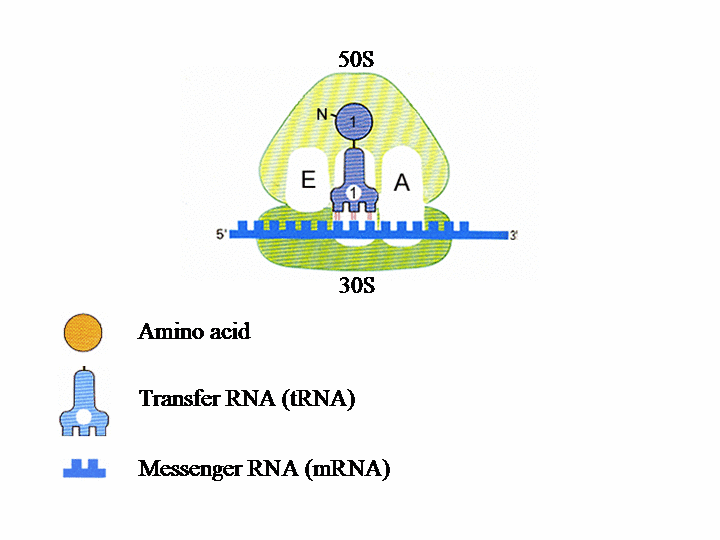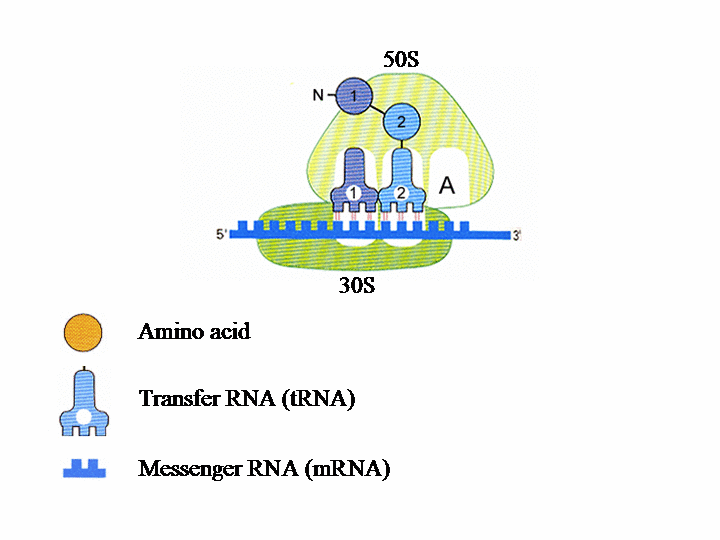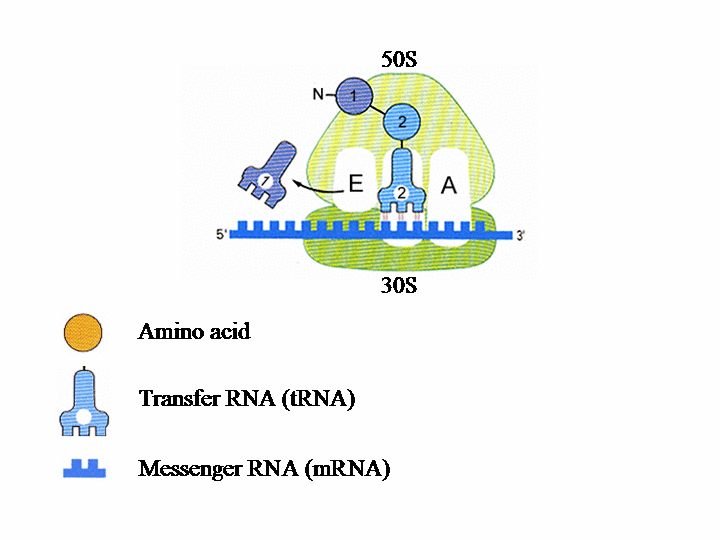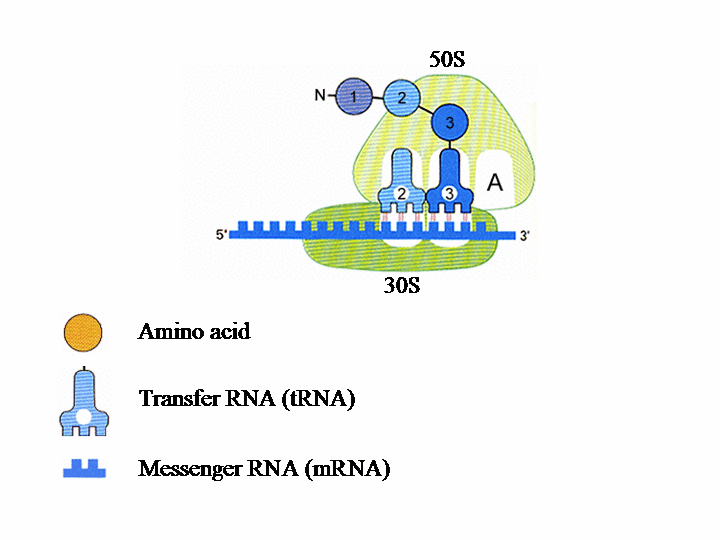
Translation of the genetic information into protein in the ribosome The ribosome consists of two subunits. The smaller, named 30S, binds to the mRNA (messenger RNA), which contains the genetic instructions needed for the synthesis of a protein. In the second subunit, 50S, the chain of amino acids that forms the protein is produced.Each tRNA (transfer RNA) “reads”, three by three, the letters forming the mRNA. Each of these groups of three “bases”, called a codon, leads to the insertion of a specific amino acid.
Each ribosomal subunit contains pockets in which the tRNA molecules bind. The P site contains tRNA1 bound to the first amino acid (AA1). tRNA2-AA2 then binds to site A and AA1 transfers from tRNA1 to AA2. tRNAs 1 and 2 and the mRNA then move with respect to the ribosome, so tRNA1 is now in site E, tRNA2 is in site P and a new codon on the mRNA is exposed at site A. tRNA3 enters site A bearing AA3 and tRNA1 is ejected from site E. This process is repeated until a “stop” codon is encountered on the mRNA
.

Atomic structure of the 30S ribosomal subunitThis high-resolution 3-D image shows the binding pockets of the 30S subunit for three antibiotics, spectinomycin, streptomycin and paromomycin. Each drug blocks a vital function of the ribosome.

Atomic structure of the entire ribosomeThe atomic structure of the entire ribosome was obtained by high-resolution X-ray crystallography. The upper part is the 50S subunit and the lower part the 30S subunit. The three tRNAs (those at sites A, P and E, see figure 1) are positioned at the interface between the two subunits.
Dr Venki RAMAKRISHNAN
Structural Studies Division
MRC Laboratory of Molecular Biology
Hills Road
UK – CAMBRIDGE, CB2 2QH
T +44 1223 402 295
F +44 1223 213 556
ramak@mrc-lmb.cam.ac.uk
http://www.mrc-lmb.cam.ac.uk/ribo/homepage/ramak/index.html
